In the context of metastatic breast cancer treatment, PET/CT imaging is considered an important diagnostic tool for assessing diseases and testing new drug therapies. Particularly, thanks to its high sensitivity, this cross-modality is widely adopted to detect metastases. However, the manual or semi-automatic segmentation of metastases, which is a preliminary step to a more precise characterization of these lesions, remains a very time-consuming operation. As a first step toward automating this process, we propose an automatic detection of liver metastases based on a combination of classic machine learning and deep learning.
Method
The study comprises 18 patients who underwent PET/CT acquisitions and from whom reference liver lesions were manually delineated by an expert radiologist using our certified Keosys medical imaging reading software. Of the 1200 contoured lesions, 67 were located in the liver.
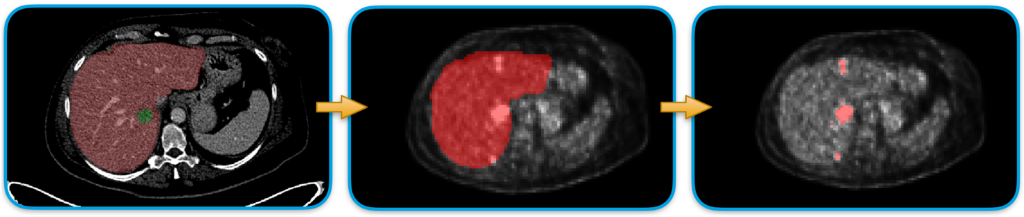
Figure 1: Liver lesion segmentation scheme.
Our designed approach consists of a series of processing steps as depicted in Fig.1. First of all, the liver segmentation from CT images is performed to delimit the searching area for the hepatic lesion detection. To do that, we trained a nn-Unet model1 on the publicly available LiTS dataset 2 composed of 130 CT cases with liver and lesions already labeled. The trained model is then used to segment the liver in our 18 patients from the Epicure project.3 A rigid registration is then applied to project the liver masks from the CT domain space into the PET domain. (See Fig. 2.)
Finally, the lesion extraction is performed by using an SVM classifier that is fed with only liver pixels characterized by an SUV greater than 2.5 and the corresponding labels (lesion or not lesion).
To cope with the limited number of cases, we split the dataset into three folds without replacement. For each fold, we performed the classifier training on 12 cases and the test on the remaining six. In this way we obtained three different models and performance estimates.
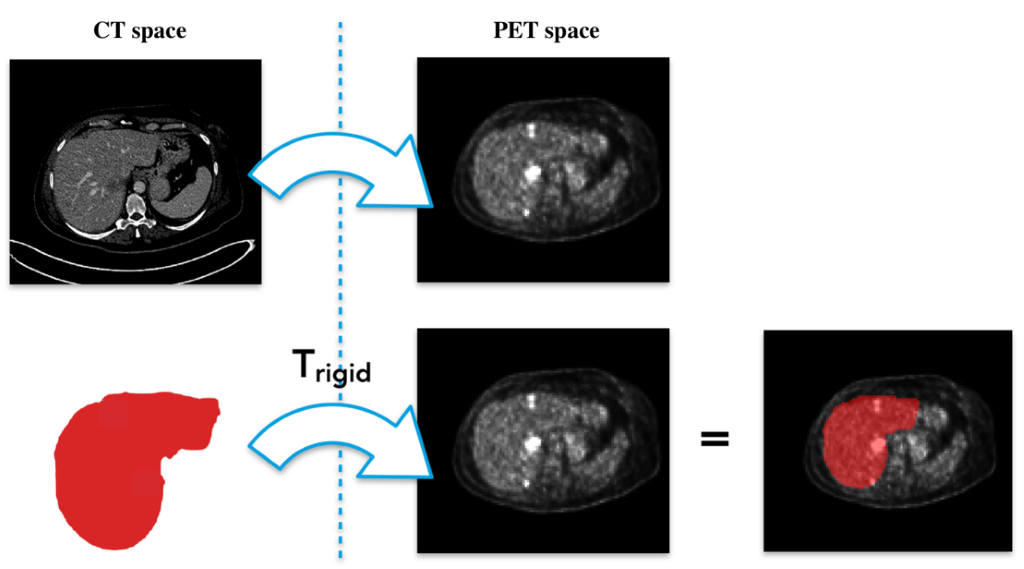
Figure 2: Rigid registration. The estimation of a rigid transformation is performed to pass from the CT space to the PET one. The estimated transformation (Trigid) is then applied on the liver mask.
Results
The liver segmentation from the CT images was evaluated with the Dice score reaching a value 0.95. The lesion segmentation quality was assessed by using pixel-wise metrics like sensitivity, ppv, and specificity. The final result was established by averaging the outcomes on the single folds obtaining an overall sensitivity of 0.47, a ppv equal to 0.80, and a specificity of 0.99. The high values of specificity and ppv highlight the model’s ability to correctly detect the presence of malignant lesions. On the other hand, the low sensibility value suggests a slight underestimation in the lesion extension process. Some results are shown in Fig. 3.
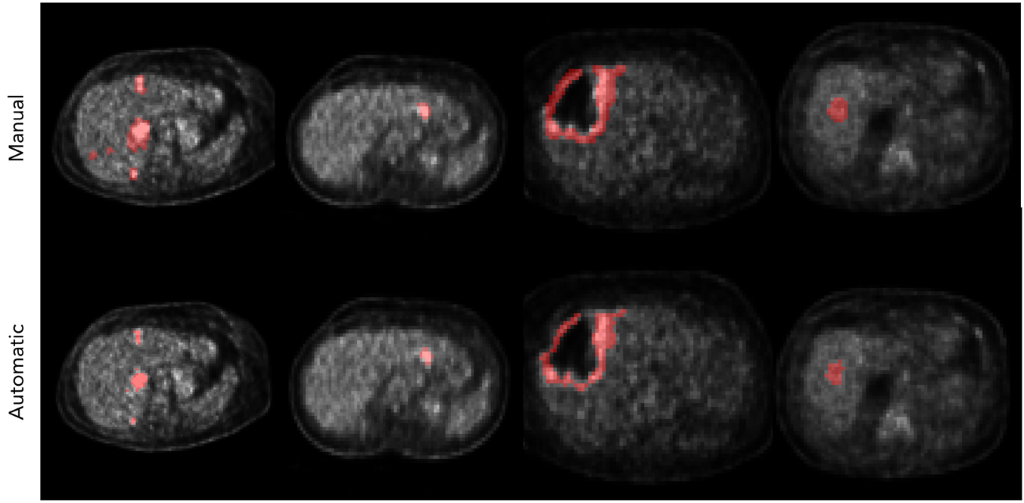
Figure 3: Comparisons between manual delineations (above) and the corresponding automatic segmentation (below)
Conclusion
In this preliminary study we presented a method to automatically detect liver lesions from PET images by combining the information coming from both PET and CT acquisitions.
References
1. Isensee, F., Petersen, J., Klein, A., Zimmerer, D., Jaeger, P. F., Kohl, S., … & Maier-Hein, K. H. (2018). nnu-net: Self-adapting framework for u-net based medical image segmentation. arXiv preprint arXiv:1809.10486.
2. Bilic, P., Christ, P. F., Vorontsov, E., Chlebus, G., Chen, H., Dou, Q., ... & Kadoury, S. (2019). The liver tumor segmentation benchmark (lits). arXiv preprint arXiv:1901.04056.
3. https://projet-epicure.fr


