Introduction
Breast cancer is the most common cancer in women and the second most frequent cancer overall. One in eight women will be diagnosed with invasive breast cancer in their lifetime. Breast cancer survival varies according to cancer staging at diagnosis. If detected early, the overall 5-years survival rate is 98% but it goes down to 27% with metastatic involvement [1]. 18FDG positron emission tomography combined with computed tomography (18FDG PET/CT) whole-body imaging is widely used for diagnosis and follow-up [2]. Based on these imaging techniques, lesion segmentation can provide information to assess a treatment effect and to adapt the treatment over time. However, manual segmentation methods are time-consuming and subject to inter and intra-observer variability.
In a first step towards automating segmentation, we compare the performances of semi-automatic traditional and deep learning-based segmentation methods.
Methods
308 metastatic lesions from 16 patients of the EPICUREseinmeta study were manually delineated by an ICO nuclear medicine physician using the Keosys viewer. Only PET images converted in SUV were used.
The SUVmax seed point was extracted from the delineations to mimic a one click user initialization, and was employed to automatically build a bounding box around each lesion. This bounding box was then used to define a region of interest on which the 5 traditional region growing-based algorithms (Nestle, 41% SUVmax, Daisne, Black and a STAPLE of the 4 previous methods) and the deep learning were applied (Fig. 1).
A 3D U-Net implementation called nnU-net was used for the deep learning. A 3-fold cross validation was done: 2/3 of the patients were used for training and 1/3 for validation. During training, the network was fed with the region of interest around the lesion and its ground truth mask. After a couple of hundred of epochs, the network parameters and weights are set to segment as well as possible the lesions. For validation, the same network with the same network parameters and weights is used to segment new lesions (Fig. 2).
Segmentation performances were evaluated calculating the Dice score between the semi-automatic segmentations and ground-truth expert manual segmentations.
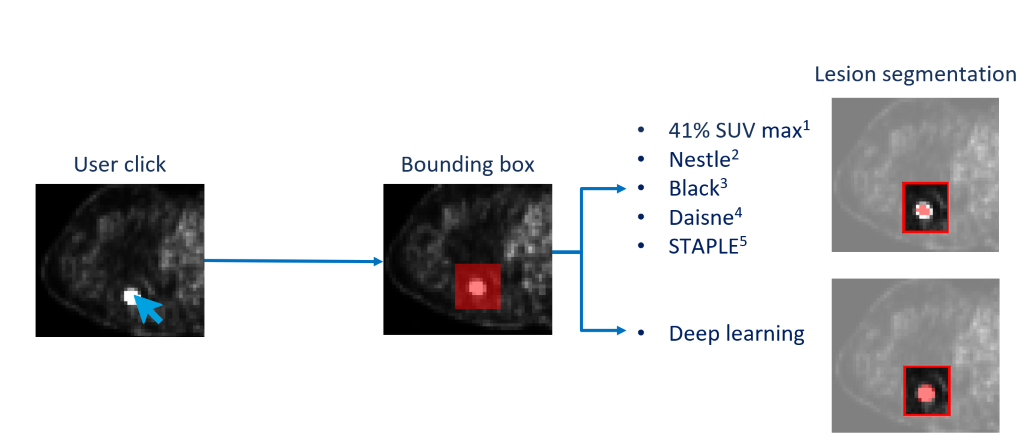
Figure 1 : Global workflow, the SUVmax is used to mimic the user click to compute the bounding box. The bounding box is used to define a region of interest on which we applied the thresholding methods and the deep learning approach.
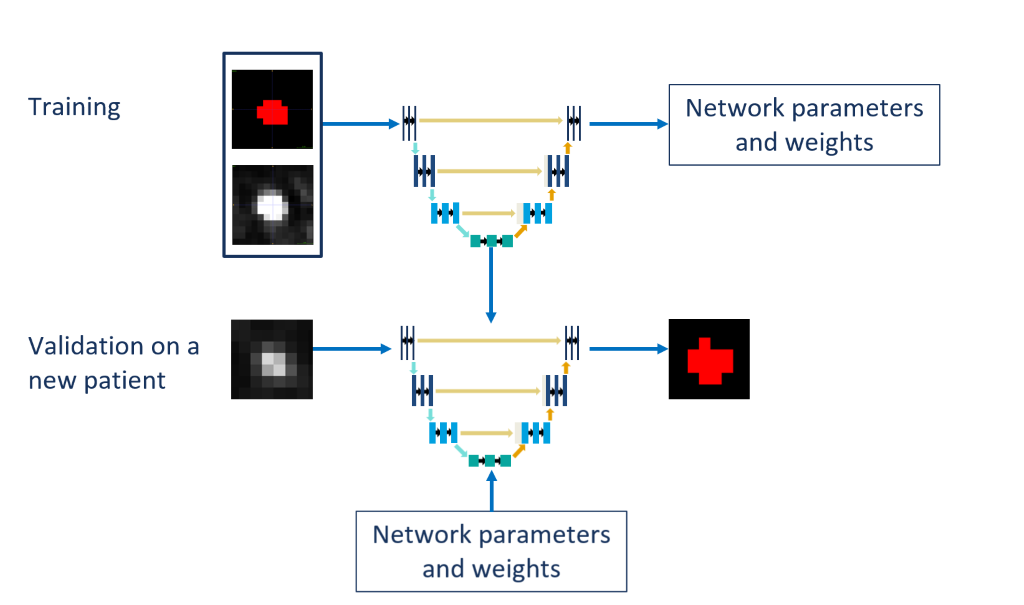
Figure 2 : During training, the network was fed with the region of interest around the lesion and its ground truth mask. After a couple of hundred of epochs, the network parameters and weights are set to segment as well as possible the lesions. For validation, the same network with the same network parameters and weights is used to segment new lesions.
Results
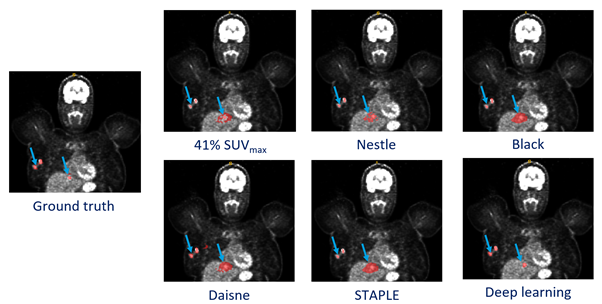
The 5 traditional methods gave very heterogeneous results depending on the location and contrast of the lesions, with a Dice score of 0.33 ± 0.24, 0.12 ± 0.07, 0.20 ± 0.15, 0.19 ± 0.13, 0.16 ± 0.14 for Nestle, Daisne, Black, 41% SUVmax and STAPLE methods, respectively. On the other hand, the deep learning approach obtained a Dice score of 0.48 ± 0.19. The Fig. 3 shows all methods used.
Conclusion
Using a deep learning approach to segment cancerous lesions semi-automatically improves the performances against traditional methods. These first results in a difficult clinical context are promising, since they could be improved, particularly by considering the anatomic site of the different lesions.