Introduction
Metastatic breast cancer requires constant monitoring, usually performed by physicians on 18F-FDG-PET/CT acquisitions. On these images, physicians interpret tumor response to treatment following standardized guidelines such as Response Evaluation Criteria in Solid Tumors (RECIST) or Position Emission Tomography Response Criteria in Solid Tumors (PERCIST). However, these guidelines tend to focus only on a selection of lesions representing tumor burden or, in the case of PERCIST, on only one lesion (the one showing the highest uptake).
Assessing the total tumor burden would be challenging and time consuming. To help physicians monitor all lesions and evaluate tumor evolution more accurately, research has notably focused on image registration methods. Indeed, if PET images acquired at different time points are precisely registered, the assessment of all lesions becomes much easier.
In the context of the Epicure project, we propose a new registration method formalized as a conventional registration approach, with a deformation field modelled by an untrained deep pyramidal network. We named our approach MIRRBA for Medical Image Registration Method Regularized by Architecture.
Method
We applied MIRRBA on our longitudinal PET images acquired for the evaluation of treatment response in patients with metastatic breast cancer. Diverging from the supervised learning paradigm, MIRRBA does not require a learning database, but only the pair of images to be registered in order to optimize the network’s parameters and provide a deformation field.
We studied the impact of different architecture configurations (16 configurations) on the deformation field on a private dataset of 110 metastatic breast cancer full-body PET images with manual segmentations of the brain, bladder, and metastatic lesions.
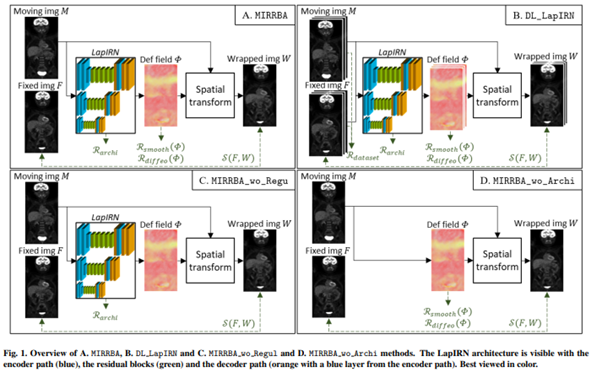
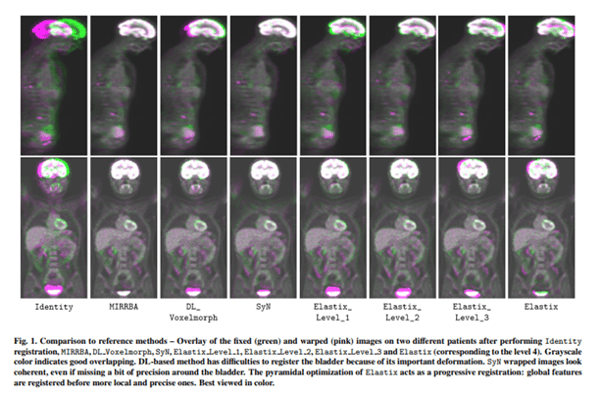
We then compared the method against several baselines: conventional iterative registration approaches (ANTs and Elastix) and supervised deep learning-based models (LapIRN and Voxelmorph). Global and local registration accuracies were evaluated using the detection rate and the Dice score respectively, while registration realism was evaluated using the Jacobian’s determinant. Moreover, we computed the ability of the different methods to shrink vanishing lesions with the disappearing rate.
Results
MIRRBA significantly improves the organ and lesion Dice scores of Voxelmorph by 6% and 52%, and of LapIRN by 5% and 65% respectively. Regarding the disappearing rate, MIRRBA more than doubles the best performing conventional approach SyNCC score.
Conclusion
Our work proposes an alternative way to bridge the performance gap between conventional and deep learning-based methods and demonstrates the regularizing power of deep architectures.
More information on this work can be found here.



